Just a reminder the next Cambridge Cheminformatics meeting is on 18 February 2026, 4-5.30pm UK time, Hybrid (at the CCDC on Union Road, Cambridge/via Zoom)
Blog

Another addition to the superb MayaChemTools OpenFECalculateRelativeBindingFreeEnergySepTop.py to calculate Relative Binding Free Energy using a Separated Topologies (SepTop) approach. The script relies on the availability of OpenFE
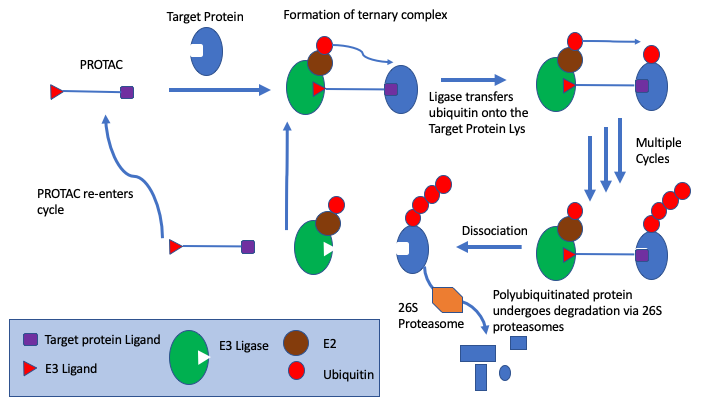
PROteolysis TArgeting Chimeras (PROTACs) technology provides an alternative to module biological function by specially using the ubiquitin proteasome system to induce degradation of the target
There is an interesting paper in Journal of Cheminformatics J Cheminform 17, 142 (2025). https://doi.org/10.1186/s13321-025-01094-1 Cheminformatics Microservice V3 aims to provide easily accessible and reproducible cheminformatics tools.
Agents are all the rage in AI/ML at the moment so it is no surprise that Apple have updated Xcode. Developers can leverage coding agents,
The popular chemistry drawing package ChemDoodle has been updated. Version 12.10.0 is a feature update for ChemDoodle 2D. Major additions include a much more powerful
CNotebook provides chemistry visualization for Jupyter Notebooks and Marimo using the OpenEye Toolkits. Import the package and your molecular data will automatically render as chemical
The next Cambridge Cheminformatics meeting is on 18 February 2026, 4-5.30pm UK time, Hybrid (at the CCDC on Union Road, Cambridge/via Zoom) More details of
MolView – SMILES Preview is a Visual Studio Code extension that renders SMILES (Simplified Molecular Input Line Entry System) strings as molecular structure diagrams on
A while back I wrote a post on the problems with maintaining scientific software, this seemed to strike a chord and I got a lot